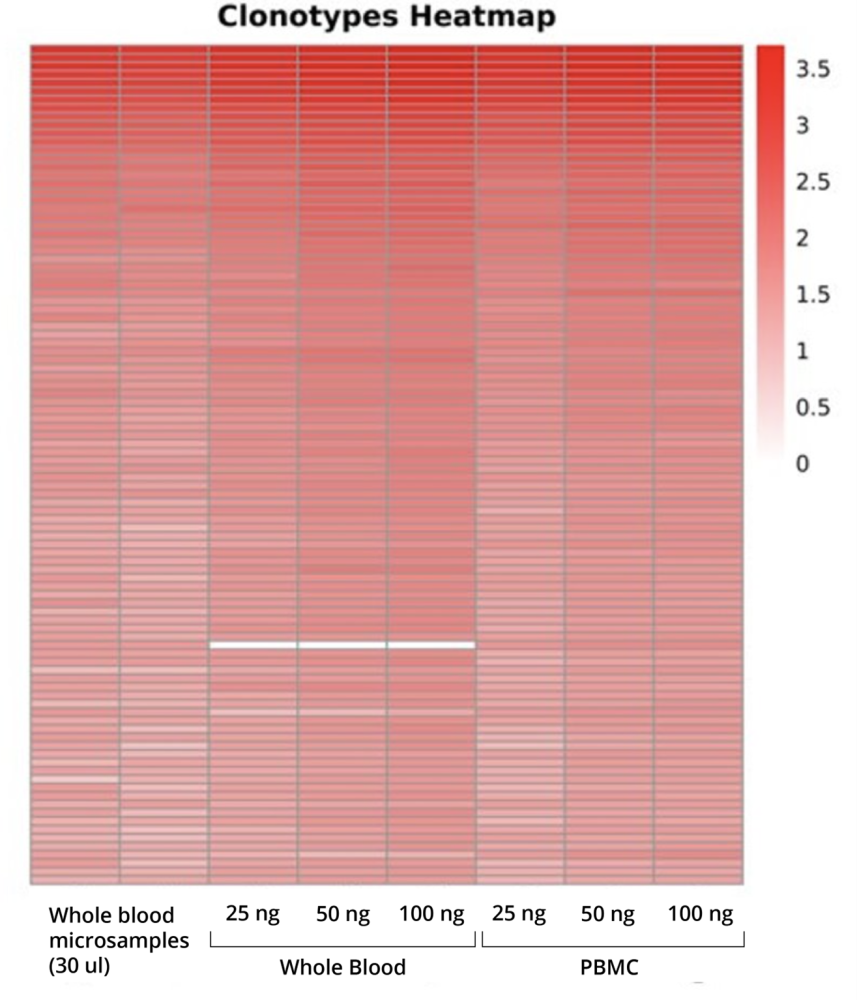
We have demonstrated that AIR RNA profiling can be done directly from whole blood samples (after purification of RNA from Tempus or PAXgene test tubes) without isolating PBMC. A comparative analysis conducted with the same amount of whole blood and PBMC samples shows that both assays detect a comparable number and similar repertoire of high- to medium-abundant clonotypes.
Based on these results, we recommend using whole blood (stabilized in Tempus) instead of PBMC for deep AIR profiling. This reduces the variation in batch effects caused by the preparation, storage, and transportation of PBMC samples and simplifies the blood collection process, especially when samples need to be collected at multiple time points [12].
Need more help with this?
Contact Us

