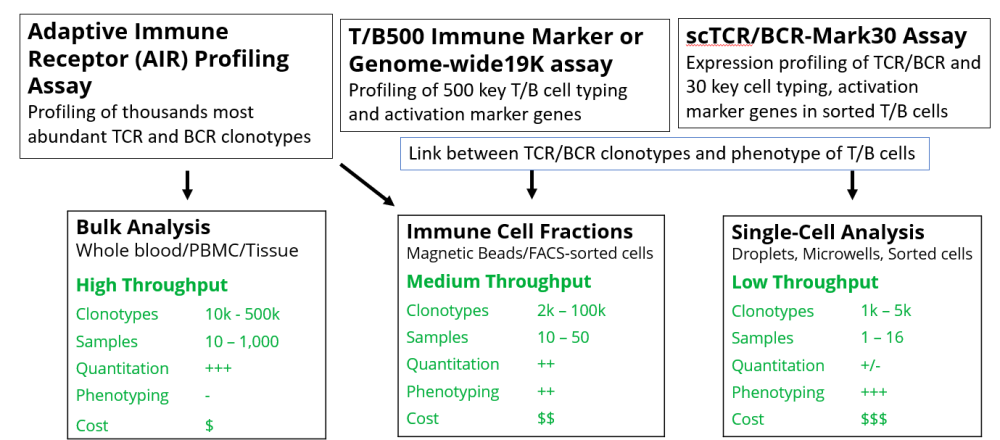
Bulk Analysis
Bulk analysis is the current front-line, high-throughput experimental strategy for comprehensive and sensitive disease-associated or treatment-associated clonotype detection. It can be performed using RNA or DNA isolated from either small or large samples of frozen or lysed blood, cell fractions, or tissues. The main advantage of bulk analysis is the simple protocol that may be conducted with standard lab equipment and scaled for hundreds of samples in 96-well plates. The cost of sequencing is reasonable, and multiple open-source software tools are available for downstream data analysis. The drawback of bulk analysis is the inability to provide information about the pairing of receptor chains and integration of CDR3 sequences with the phenotype of T and B cells for each clonotype. Although cell-type deconvolution algorithms applied to RNA-seq data can potentially link cell subtypes with the most abundant clonotypes, in practice, the application of this strategy is currently limited [10].
Single-cell Analysis
Single-cell immune receptor profiling is a revolutionary approach that allows investigators to combine clonotype repertoire with paired-chain information and the phenotype of cells (e.g., cell subtype). Single-cell immune receptor profiling assays can be performed at low throughput (96-384 cells) using sorted cells in 96-well plates (e.g., AIR RNA assay) or at medium throughput (1,000-5,000 cells) using microwell arrays or droplet microfluidics-based technologies [1-5, 9]. However, single-cell immune receptor profiling assays are more complicated and expensive (for both reagents and sequencing) than bulk assays. They typically require live cells, which is not practical for many applications. Furthermore, single-cell immune receptor repertoire analysis can only profile (in a non-quantitative fashion) the most abundant clonotypes due to the low number of cells analyzed in the assay.
FACS Sorted Immune cells or fractions
To address the limitations of single-cell analysis, a middle strategy is to profile immune cell fractions (e.g., T/B naive, memory, effector, exhausted cells) isolated by antibody-loaded magnetic beads or FACS-sorting from blood or tissue samples using conventional cell-typing-specific antibodies. FACS-purified immune cell fractions (e.g., 5K-50K cells) can be used directly without purification for DriverMap AIR RNA and a DriverMap T/B immune marker assays developed for expression profiling the 500 top T- and B-subtyping and activation marker genes. By combining AIR profiling with expression profiling of key immunity genes in the same sorted cells, you can link the clonotype information with the cellular phenotype of cells. [13,18]
Need more help with this?
Contact Us

