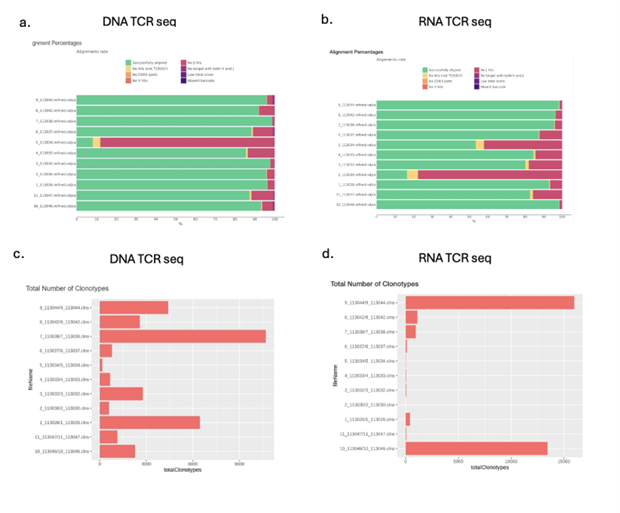
FFPE (Formalin-Fixed, Paraffin-Embedded) samples are a common way to preserve tissue. While they allow for long-term archiving of specimens, the chemical cross-linking and partial degradation from formalin can make it challenging to extract high-quality RNA. Successful TCR/BCR profiling from FFPE tissues depends not only on the content of lymphocyte cells but also on the quality of the extracted RNA.
While FFPE AIR-RNA profiling is possible to do, it requires a RIN Score > 5, or using RNA with at least 20% of fragments above 300-n. For FFPE RNA input amounts, we recommend maximizing the RNA input amount (up to 1-2ug, if possible) to increase the sensitivity of the AIR assay. In our experience, DNA is more stable and provides more reliable data when working with FFPE samples. However, due to the higher copy number of RNA vs DNA molecules per cell, using RNA can give more sensitive profiling of immune receptor clonotypes than DNA if RNA is not significantly degraded. For the best results, we recommend purifying both RNA and DNA and running both AIR-RNA and AIR-DNA assays in parallel (see Fig. 20 below).
When working with FFPE samples, it is critical to recognize that the content of B and T cells could be very different. Therefore, we recommend running AIR-TCR and AIR-BCR assays separately based on the content of T and B cells (e.g. using H&E staining). In addition, FFPE-DNA and impurities in DNA samples can inhibit DNA polymerase activity for AIR-DNA profiling. Therefore, to identify the optimal input amount under these conditions, we suggest testing (for at least one FFPE-DNA sample) a range of DNA inputs (e.g., 0.5 µg, 1 µg, 2 µg, 5 µg, for 50-ul AIR primer extension step), though 1-2 µg has generally shown optimal sensitivity without inhibition of DNA polymerase activity. If the amount of purified FFPE-DNA is 5-20ug, you could consider running replicate samples (e.g., 2 ug in 50-ul primer extension step) to increase the sensitivity of AIR profiling.
Need more help with this?
Contact Us

