Please refer to the AIR Technology Guide for recommendations for experimental design, sample-input amounts, and assay design.
Assay:
Q1: What is included in the AIR profiling kit?
A1: The kit includes all the necessary reagents for reverse transcription, amplification, and primers for next-generation sequencing.
Q2: What is the difference between TCR/BCR CDR3 RNA kits for human and mouse?
A2: The TCR/BCR gene-specific primers for the human and mouse kits are different but all other reagents and protocols are the same.
Q3: Does the AIR profiling assay have Unique Molecular Identifiers (UMIs)?
A3: Cellecta’s DriverMap AIR RNA and AIR DNA profiling assays do not have UMIs. The AIR RNA and AIR DNA profiling assays incorporate a redundant set of Validator Barcodes (VBCs) which enable easy identification of and correction for amplification and sequencing biases and error. The VBCs also provide a reference for normalization and copy number quantitation of amplified RNA or DNA molecules to facilitate detection of low-abundant clonotypes.
Q4: What is the difference between AIR RNA CDR3 and Full-length Receptor Kit?
A4: The CDR3 kit amplifies approximately 250-300 bp of the CDR3 region, whereas the full-length kit amplifies the full variable region ~600 bp (CDR1-CDR2-CDR3). For more information on sequencing instructions and applications for the two kits, please visit the AIR-RNA webpage to view the table Comparison of CDR3 vs Full-Length Variable Region Repertoire Profiling.
Q5: What sample types are compatible with the assay?
A5: Our assay is compatible with various sample types, including RNA or/and DNA samples isolated from whole blood, PBMC, immune cell fractions, tissue/cancer biopsies, or FFPE tissue samples. For more information on sample types and preparation guidelines, please refer to the Technology Guide; Applications of AIR profiling.
Q6: What is the recommended protocol for profiling a small number of cells, such as sorted cells or fresh frozen tissue?
A6: For a small number of cells (e.g., 5,000-50,000 cells), e.g., isolated by FACS, tissue or blood microsamples, we recommend using the Cellecta DirectCell™ Protocol which profiles directly from the cell lysate without the need to extract total RNA or DNA from the sample.
Q7: What is the optimal input amount for DNA as a starting material?
A7: To determine the optimal input amount for your samples, we recommend testing amplification efficiency using different starting amounts of DNA, e.g. 0.5 µg, 1 µg, 2 µg, 5 µg. Some samples such as FFPE blocks, or tumor biopsies contain impurities that inhibit the activity of DNA polymerase, so a maximum of 2 µg is recommended.
If you have enough input amount, we recommend running samples in triplicates, e.g. using 2×3=6 µg of DNA for each sample to get better quantitative data, as shown in Technology Guide Section 2.1, Fig. 9, which lists the number of clonotypes you can obtain from different amounts of input DNA material.
Q8: What is the better choice of starting material for FFPE samples, DNA or RNA?
A8: In our experience, DNA, due to its stability, generally provides more robust data with FFPE samples. However, RNA can lead to a more sensitive profiling of clonotypes provided that the RNA is not significantly degraded. For FFPE AIR-RNA profiling, we recommend using RNA with a RIN Score > 5, or RNA with at least 20% of fragments above 300-n. For best results, we recommend purifying both RNA and DNA from FFPE samples and running both AIR-RNA and AIR-DNA assays.
Q9: Can less than 500 ng of DNA be used for the AIR DNA Profiling Kit?
A9: As shown in Technology Guide Section 2.1, Fig. 9, profiling with 500 ng or lower is possible, but the number of detected clonotypes will be reduced. Furthermore, only top abundant clonotypes (with clonotype-specific cells numbering more than 10) could be quantitated.
Sequencing:
Q10: What is the recommended number of reads for the AIR protocol?
A10: For sequencing, we recommend around 5-10 million reads per sample, depending on the number of samples in one flow cell and the required sensitivity. More information about sequencing can be found in the User Manuals’ NGS Prep and Sequencing Section.
Q11: How much PhiX is needed for the sequencing run?
A11: We recommend adding 15% of PhiX to the library (anywhere in the range 10-20% is acceptable).
Q12: Can AIR RNA and AIR DNA samples be sequenced together in the same lane?
A12: Yes, AIR-RNA and AIR-DNA samples can be combined for NGS, but due to differences in amplicon sizes (approximately 350 bp for RNA vs. approximately 250 bp for DNA), we suggest mixing AIR-RNA with AIR-DNA at a 1.5:1 ng ratio (e.g., 150 ng of AIR-RNA with 100 ng of AIR-DNA). Be sure to use the different indexing primers for RNA and DNA samples. NGS sequencing primers are the same for both AIR-RNA and AIR-DNA.
Q13: Which instruments can be used for CDR3 or full-length receptor sequencing?
A13: For most applications, we recommend using the Illumina NextSeq 2000 instrument with SBS-Leap sequencing chemistry kits due to low NGS cost and wide range of flow cells (between 100M to 1.8B reads for both 300-cycle and 600-cycle Paired-End reads). The NextSeq 500/550 could still could be used for CDR3 profiling, but the cost of sequencing is higher than when using the NextSeq 2000. The MiSeq instrument allows sequencing for the 600 cycles (full-length receptor sequence) but its scale is limited to 5-10 samples. Our kits use the reverse complement workflow, so for MiSeq, you would need to use the protocol with one index primer and the V2/V3 Kits. To optimize your sequencing protocol, please contact techsupport@illumina.com.
Q14: Can you sequence DriverMap AIR and DriverMap Human Genome-Wide Targeted RNA-Seq Expression (EXP) Profiling Kits in the same flow cell?
A14: Yes, the custom sequencing primers (Reverse and Forward SeqDNA and SeqIND primers) are the same in the DriverMap AIR and DriverMap Human Genome Wide Expression Profiling Kits. Please note that AIR and Expression profiling samples need to be amplified with different indexing primers to be sequenced in the same flow cell.
Q15: Can the libraries be sequenced on the MiSeq? Do you have any recommendations?
A15: Yes, the AIR library can be sequenced on the MiSeq instrument. Our kits use the reverse complement workflow, so only the V2/V3 kits are compatible with the MiSeq and you would need to use the protocol with one index primer. Attached is the Illumina Guide for Indexed Sequencing Overview. To optimize your sequencing protocol please contact techsupport@illumina.com.
Q16: Is it recommended to do sequencing on the NovaSeq 6000 or NovaSeq X Platform?
A16: As NGS with NovaSeq instruments provides a minimum of 8B reads per flow cell, NovaSeq instruments could be used for NGS analysis of AIR samples that are done on a large scale (i.e.> 500 samples).
Q17: Can sequencing be performed on NovaSeq 6000?
A17: Yes, it is possible to use two custom indexing primers with the NovaSeq 6000 Reagent Kit v1.5, as the NovaSeq 6000 machine works with a mixture of Ind1 and Ind2 sequencing primers from one well. The recommended approach is to spike Cellecta primers in the wells with Illumina primers, as described in the Spiking custom primers into the Illumina sequencing primers | Illumina Knowledge. If you prefer not to spike in the primers, you can use the custom primers protocol described in the NovaSeq Series Custom Primers Guide (illumina.com).
We have a customer that used the SP flow cell and loaded 2nM with the protocol with 148-10-10 -148 cycles as follows:
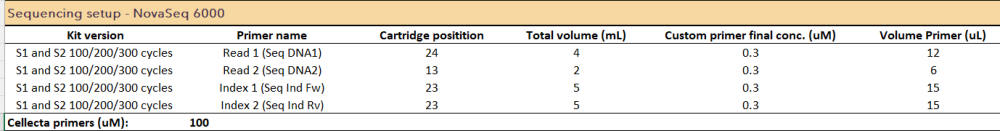
Please note that Cellecta has not internally validated this protocol, so to optimize your sequencing protocol please contact techsupport@illumina.com.
Q18: What are the recommended concentration and sequencing primer setup for NovaSeq 6000 kits?
A18: For NovaSeq 6000, we generally recommend starting at 100 pM (same as PhiX) if the library type loading concentration is unknown, then optimizing in 50-100 pM increments based on results. Here is Illumina resource for determining optimal loading based on PF and occupancy. To optimize your sequencing protocol please contact techsupport@illumina.com.
Q19: Can sequencing be performed on NovaSeq X?
A19: Yes, our kits use Dual DNA/RNA UDP Indexes and follow the reverse-complement workflow. For loading on the NovaSeq X, follow Illumina’s protocol for NovaSeq X Custom Primers Protocol. To optimize your sequencing protocol please contact techsupport@illumina.com.
Data Analysis:
Q20: How can I analyze the data for TCR and BCR analysis?
A20: We recommend using the MiXCR software tool (MiLaboratories) for alignment and AIR repertoire data analysis. Here are the options for our customers:
● Academic customers: Academics may use the free, user-intuitive MiXCR software available for download on Platforma.bio.
● Industry customers: MiLaboratories offers a free trial period to test our kits on Platforma.bio or MiXCR command line version, starting with a 1-month free trial onwards. MiXCR packages include special presets for AIR kit analysis, the complete workflow can be set up with the name of the AIR Kit eg: MiXCR Guides Cellecta Kit
● Cellecta bioinformatics services: Additionally, Cellecta offers bioinformatics services, including running the MiXCR pipeline and providing output reports. Custom analysis can also be performed upon request. Here is an example report: 2024-02-13 Final Report. Request a quote or more information at quotes@cellecta.com.
Other bioinformatics tools/frameworks are also available to analyze immune repertoires as follows:
| Tools | Published year | Aligner | Website |
|---|---|---|---|
| IMGT/HighV-QUEST | 2012 | GPA (NW) | https://www.imgt.org/IMGTindex/IMGTHighV-QUEST.php |
| IgBLAST | 2013 | BLAST | https://www.ncbi.nlm.nih.gov/igblast/ |
| IMSEQ | 2015 | SCF matching | https://www.imtools.org/ |
| LymAnalyzer | 2015 | Fast-tag | https://sourceforge.net/projects/lymanalyzer/ |
Q21: How do I obtain a MiXCR license?
A21: For an academic license go to https://licensing.milaboratories.com/, for a business license email licensing@milaboratories.com and provide a Cellecta kit reference.
Need more help with this?
Contact Us

