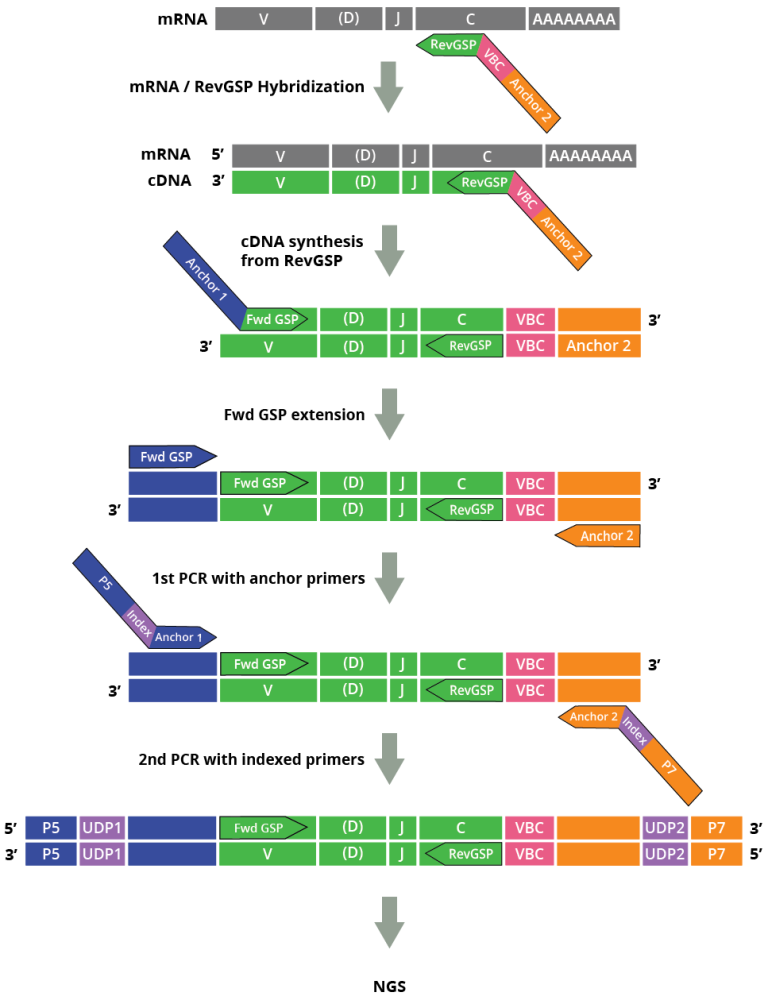
The DriverMap™ AIR RNA profiling assay is a next-generation targeted, multiplex RT-PCR technology designed for bulk expression profiling of all CDR3 TCR (TRA, TRB, TRD, TRG) and/or BCR (IGH, IGK, and IGL) clonotypes in a single reaction using RNA as the starting material. The assay is based on 315 forward primers designed for the FR3 variable region and 19 reverse primers designed for the conservative C-region of TCR/BCR mRNAs. These primers have been experimentally validated and optimized for the best performance. An additional set of primers was designed for the FR1 region, which allows (in combination with C region primers) the user to amplify full-length CDR1-CDR2-CDR3 V regions of all seven immune receptor genes (Fig. 1).
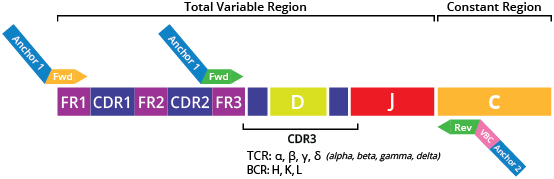
The AIR RNA assay profiles all functional or productive isoforms and excludes non-functional pseudogenes and ORFs (as defined by the IMGT database https://www.imgt.org/IMGTrepertoire/) (Fig. 2). The AIR RNA assay is designed to profile the TCR and BCR repertoire together (preferred strategy) or separately (if necessary).
Need more help with this?
Contact Us

