The DriverMap™ AIR DNA profiling assay is a multiplex PCR assay that uses the same set of 315 forward primers designed for the FR3 region and 100 reverse primers designed for the J region of seven immune receptor genes (Fig. 3).
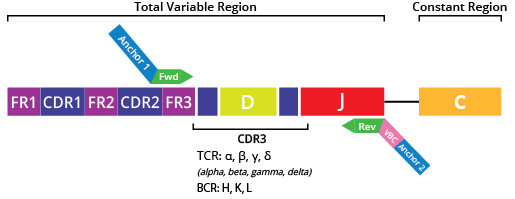
Similar to the AIR-RNA assay, the AIR-DNA assay allows you to amplify all these seven chains encoded by the TCR and BCR genes. As there is a significant difference in the number of T and B cells present in biological samples, the preferred strategy for balanced immune receptor profiling from DNA is to amplify the TCR and BCR chains in separate reactions. (Fig. 4).

Last modified:
21 February 2025
Need more help with this?
Contact Us

