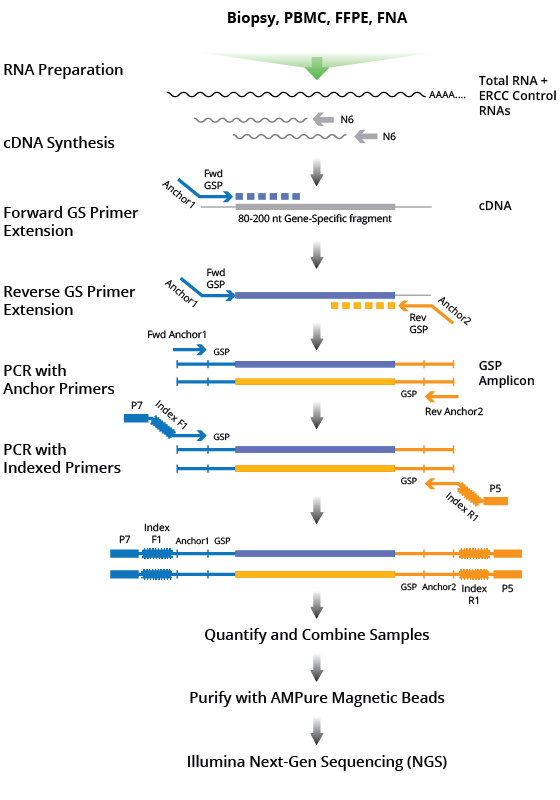
Cellecta’s DriverMap™ Targeted Expression Profiling Kits provide reagents for targeted RNA expression profiling, directly from total RNA samples, or mRNA captured in oligo-dT TurboCapture microtiter plates using the DirectCell Protocol. No preliminary poly(A) selection or ribosomal/mitochondrial/globin RNA depletion is required.
The protocol utilizes a mix of empirically validated RT-PCR primers optimized for a single-tube assay protocol that prepares NGS libraries compatible with Illumina Next-Gen Sequencing (NGS) systems (NextSeq, HiSeq, or MiSeq). With the DriverMap Assay, RNA samples can be processed for sequencing in approximately 8 hours time with just around 2 hours hands-on time.
Please follow these general guidelines when running the DriverMap Assay:
- Keep enzyme components on ice during use. All other components, including primer pools, may be placed at room temperature. Thaw components, gently vortex, and spin down before use. Please be sure to dissolve any precipitate if visible.
- Use Good Laboratory Practices to minimize cross-contamination of products. If possible, perform the first part of the procedure (from cDNA synthesis through First PCR setup) in a location set aside for RNA work, and use a set of equipment, pipettes, test tubes, and other consumables dedicated for work involving RNA. Then, run the PCR amplification reactions in a separate lab or area designated for PCR. Always change pipette tips for adding components to new samples.
- To minimize the hands-on time and mistakes in liquid deposition, we recommend to run the assay in a 96-well plate using and 8-channel (or 12-channel) pipet. After adding all the necessary reagents, seal the plate well with Clear Adhesive Film and use a Compression Pad to minimize evaporation from experimental samples. Only use each Clear Adhesive Film once. Do not reuse them.
- Do not reuse clear adhesive films to seal 96-well plates.
- Pipet viscous enzyme solutions slowly. After adding them to the reaction mix, ensure complete mixing by vortexing or pipetting up and down several times.
- For several steps in the procedure, we recommend making a Master Mix by combining adequate amounts of the key reagents into a single tube, and then aliquoting the appropriate portion from this combined mix into each sample. This approach helps ensure a consistent amount of each reagent is added to all samples. For these Master Mix reactions, first calculate the volume of each component you need for all the reactions you are running. Then, add 5% to each volume to cover pipetting variance, and pool the appropriate volumes of reagents together into a single tube or well. Mix the pooled reagents, then pipette the required portion into each sample. If you are using a multichannel pipette, aliquot the Master Mix in a tube strip first, then pipette from this strip into each sample.
Need more help with this?
Contact Us