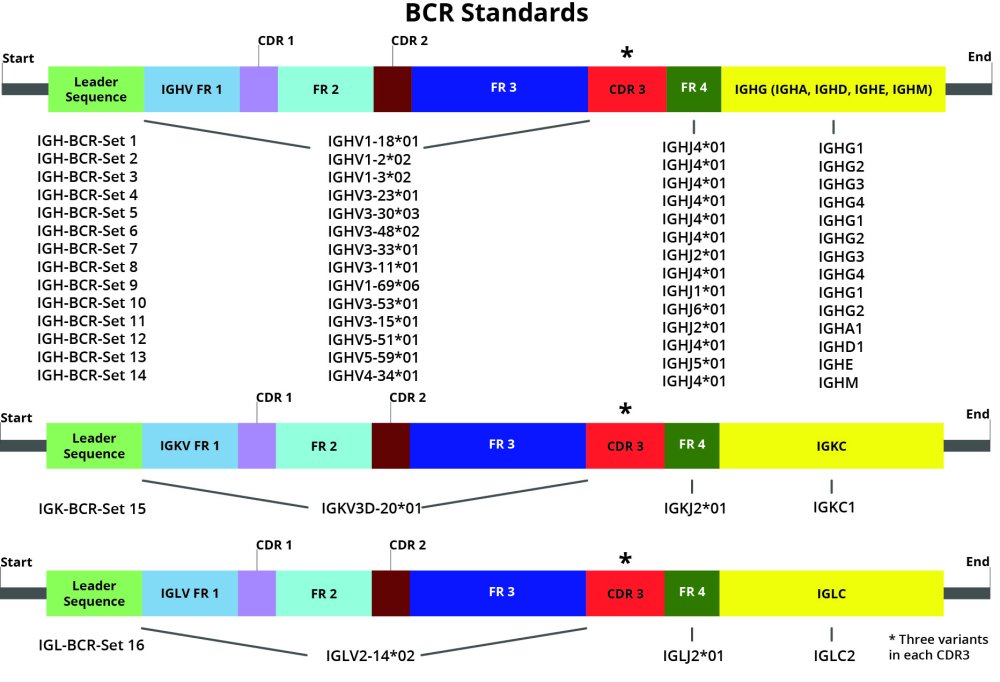
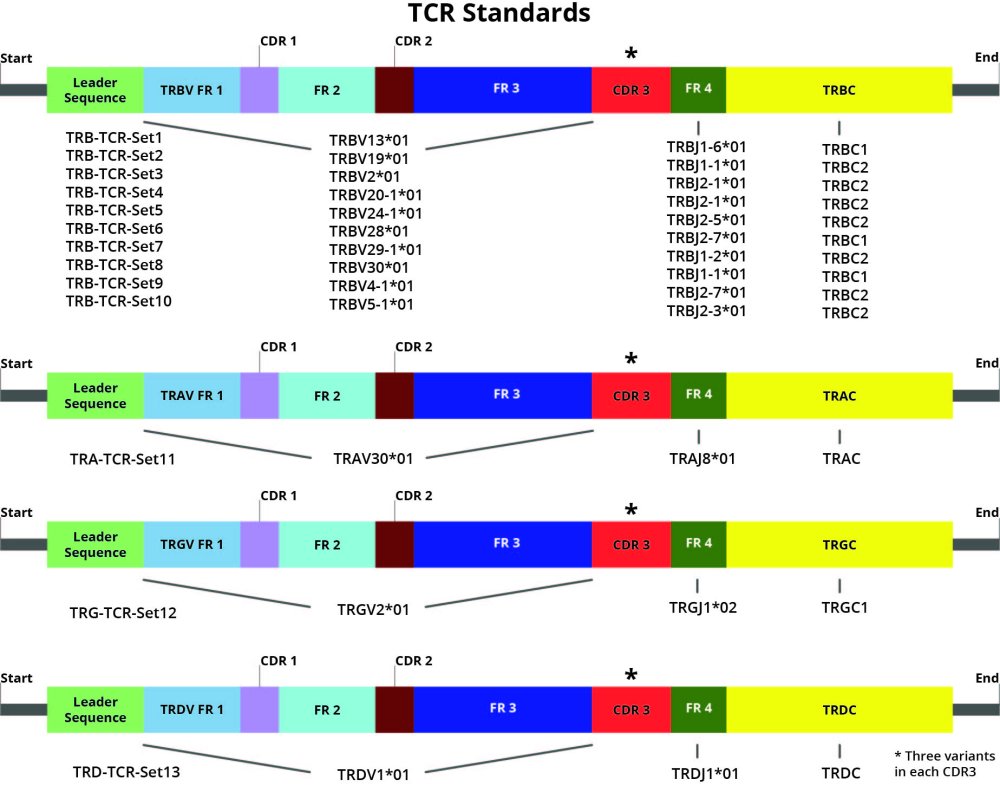
DriverMap AIR Spike-in controls are synthetic constructs for TCR or BCR in RNA format. They are designed with several unique combinations of V(D)JC genes (isoforms), each construct having a unique CDR3 sequence. Each isoform is represented by three different constructs/variants (a, b, and c) with the same V(D)JC structure but different CDR3 sequences.
AIR Spike-in Controls are available in two formats:
- Premixed Control Mixes: Premade mixes of 48 BCR and 39 TCR synthetic RNA constructs that contain all 16 BCR V(D)JC Isoform with 3 CDR3 variants for each isoform (i.e., 48 synthetic BCR constructs) or all 13 TCR V(D)JC isoforms with 3 CDR3 variants for each isoform (i.e., 39 TCR constructs), respectively. In the Premixed Controls, the three CD3 variants from each V(D)JC Isoform Pool are mixed in a 16:4:1 ratio as described in Appendix A, so the three CD3 variants of each isoform are present in three discrete concentrations 4000, 1000 and 250 molecules per microliter.
- Triplex Isoform Pools: Set of the 16 BCR and 13 TCR pools have three synthetic RNA constructs with the same V(D)JC isoform receptor structure, but each construct has a different unique CD3 sequence. The sequences of the three CD3 variant constructs within each Pool differ from each other by 3 unique single-nucleotide mutations, and the three variants are mixed at a 1:1:1 ratio with 2,500 molecules/µl each. Please refer to the Appendix section for specific TCR and BCR construct sequences, concentrations, and ratios for Triplex Isoform Pools and AIR Control Mix.
DriverMap AIR Spike-in Control Mix Kit contains TCR/BCR isoform pools or mixes at different concentrations calculated based on the A260 OD measurement of individual RNAs. The “theoretical” concentration is the number of molecules per microliter. However, based on our experimental validation results using digital-droplet PCR (ddPCR), this theoretical concentration is approximately 2x higher than the concentration measured by ddPCR. We observed that the number of construct-specific molecules detected in the AIR-RNA profiling experiment by data alignment software such as MiXCR (MiLabs Inc.) is approximately 20-60% of the “theoretical” number of molecules spiked in the RNA sample.
For example, if you spike in 5,000 “theoretical” control RNA molecules in an experimental RNA sample and run the DriverMap AIR-RNA assay, it is expected to reveal approximately 1,000-3,000 molecules at the NGS-data analysis step. The number of detected molecules for each V(D)JC-specific isoform control construct can be different (usually +/- 2-fold), which reflects differences in the performance of PCR primers used in AIR assay. The difference between theoretical and measured concentration can be attributed to losing control molecules at the library prep stage, such as purification with SPRIselect magnetic beads, pipetting, and enzymatic steps, and contamination of short RNA products in the final control RNAs.
Need more help with this?
Contact Us

