In this step, the yield of products from the Index Primer PCR Reaction—the Amplified Indexed Libraries of transcripts from each sorted plate sample—is analyzed, measured, adjusted if necessary, and then pooled in equimolar amounts for sequencing.
Perform QC and Quantify Amplified Indexed Libraries
- Analyze the Amplified Indexed Libraries using one of the following methods:
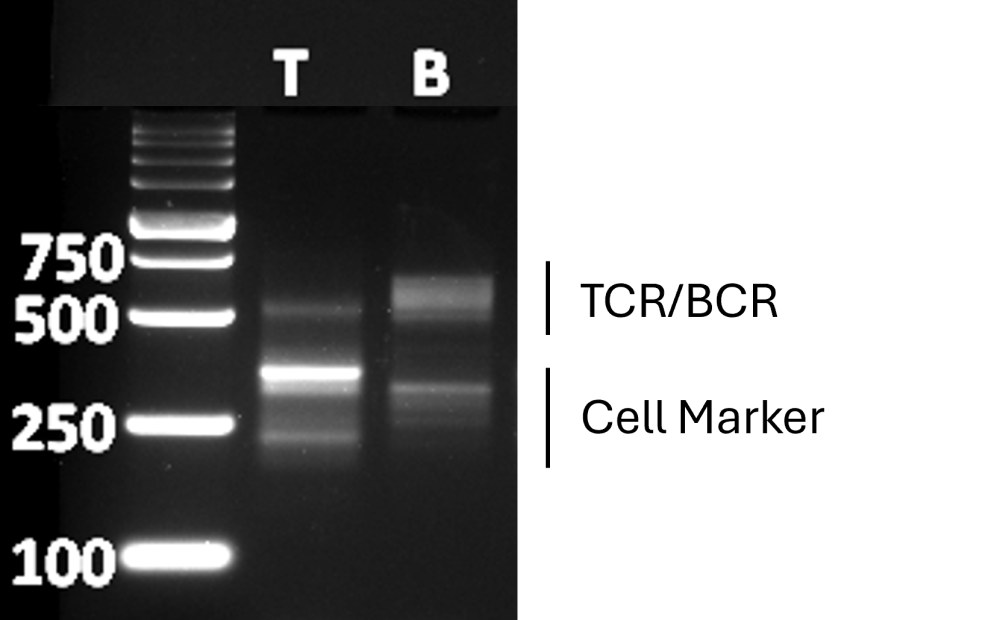
- Standard Method: Separate 5 µl of Amplified Indexed Libraries on a 3% agarose-TAE gel and analyze the size distribution of NGS probes by UV transilluminator. See Fig. 3 below for the expected results of the amplified library generated from scAIR positive control RNA (1 plate) after 2nd PCR step with indexed primers.
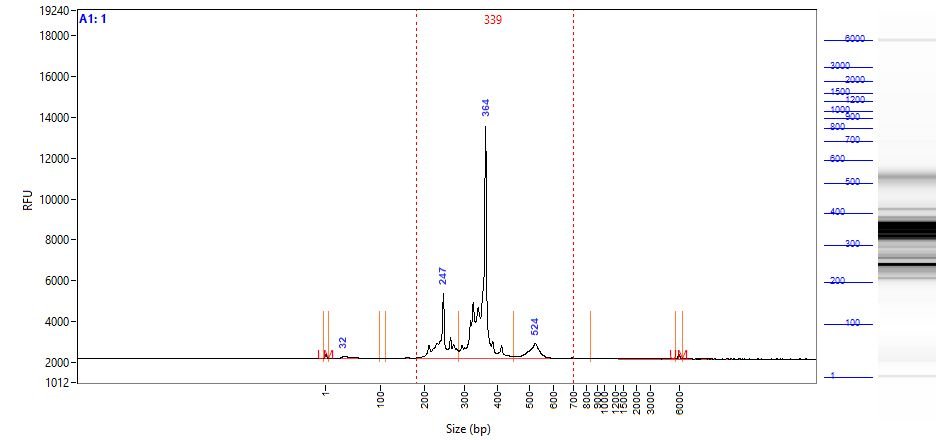
- Alternative Method: Analyze 1 µl of each of the Amplified Indexed Libraries on either an Agilent Bioanalyzer with the Agilent High Sensitivity DNA Kit (Cat.# 5067-4626) or Fragment Analyzer using the High Sensitivity NGS Analysis Kit (Cat.# DNF-473-1000) using the manufacturer’s protocol. Fig. 4 below shows the results of the analysis of amplified 2nd PCR products using Fragment Analyzer. For the scAIR TCR-Mark36 assay, T cell marker amplicons generate the smear with several bright bands in the 220-420 bp range and CDR1-2-3 TCR amplicons have a size of approximately 550 bp. For the scAIR BCR-Mark30 assay, B cell marker amplicons generate the smear with several bright bands in the 250-420 bp range and CDR1-2-3 BCR amplicons have a size of approximately 600 bp.
- Standard Method: Separate 5 µl of Amplified Indexed Libraries on a 3% agarose-TAE gel and analyze the size distribution of NGS probes by UV transilluminator. See Fig. 3 below for the expected results of the amplified library generated from scAIR positive control RNA (1 plate) after 2nd PCR step with indexed primers.
- Adjust the Second PCR cycle number (optional step)
The yield of amplified products after 2nd amplification step significantly depends on the transcriptional activity of sorted cells. For example, activated T or B cells are more transcriptionally active than naïve cells. Amplified products generated from Positive Control RNA (included in the kit) could be a useful reference for the analysis of PCR products and adjustment/troubleshooting of the scAIR protocol. If the analysis of amplified products didn’t reveal any visible yield of expected PCR products, run the sample with the amplified indexed library for an additional 3-5 PCR cycles and repeat the analysis. Optimize the number of cycles in 2nd PCR step to get visible PCR products in all experimental samples (generated from different plates). It is not necessary to achieve the same yield of PCR products for different amplified indexed libraries, differences in the yield between samples will be adjusted when the samples are combined in the next step.
- Quantify amplified indexed libraries using Fragment Analyzer/Bioanalyzer: Analyze yields of the Amplified Indexed Libraries using Fragment Analyzer/Bioanalyzer software and combine different Amplified Indexed Library products (from all replicate samples) together in equal amounts using quantitation gates set-up between 200-600bp (for scAIR-TCR-Mark36) or 200-700bp (for scAIR-BCR-Mark30). Please refer to Section 8 for recommendations on how many samples (replicates) can be combined together for different NGS flow cells. If PCR primers complicate data analysis, we recommend removing PCR primers by incubation of amplified products for each sample with Primer Removal Enzyme (see step below) and repeat analysis/quantification of Amplified Indexed Libraries in Bioanalyzer or Fragment Analyzer.

- Remove excess primers from the pooled Amplified Indexed Library by adding 2 µl of Primer Removal Enzyme to the combined sample (e.g., 100 µl), then incubate at 37°C for 30 minutes.
Last modified:
25 March 2025
Need more help with this?
Contact Us

