In this step, the yield of products from the Index Primer PCR Reaction—the Amplified Indexed Libraries of transcripts from each sample—are analyzed (adjusted if necessary), measured, and then pooled in equimolar amounts for sequencing.
QC and Quantify Amplified Indexed Libraries
- Analyze the Amplified Indexed Libraries using one of the following methods:
- Standard Method: Separate 5 μl of Amplified Indexed Libraries on a 3% agarose-TAE gel and analyze the size distribution of NGS probes by UV transilluminator. To minimize the sample number, you could run only one sample from each triplicate set. See below for the expected results of amplified libraries generated from good-quality whole-blood RNA samples. For the AIR TCR-BCR assay, the smear should be around 600 bp.
- Alternative Method: Analyze 1 µl of each of the Amplified Indexed Libraries on either an Agilent Bioanalyzer with the Agilent High Sensitivity DNA Kit (Cat.# 5067-4626) or Fragment Analyzer using the High Sensitivity NGS Analysis Kit (Cat.# DNF-473-1000) using the manufacturer’s protocol.

- Analyze yields of the Amplified Indexed Libraries. The yield should be roughly the same for all experimental samples within +/- 2-3-fold levels and similar to the Positive Control RNA sample. A negative control sample should not generate any significant yield of amplified products. If some samples show a significantly lower yield of amplification products, it could indicate differences in the amount, quality of RNA, or content of TCR/BCR mRNAs used in AIR assay. For the experimental RNA samples with a significantly lower yield of PCR product (e.g., >5-10-fold) than other samples or positive control RNA, you can re-run the lower yield samples in the thermal cycler for 2-5 additional cycles (see Note below). After cycling, quantify the products again relative to the Positive Control RNA. Note: Do not include the Positive Control RNA sample in additional cycles. Remove the positive control sample from the plate and keep it as a reference to assess the number of PCR cycles.
Remove Excess PCR Primers and Combine Samples
- Remove excess primers from the completed PCR reactions by adding 1 µl of Primer Removal Enzyme to each of the Amplified Indexed Libraries and the Negative Control sample, then incubate at 37°C for 30 minutes.
- To ensure accurate quantification for sequencing, you should repeat the quantification procedure of the Amplified Indexed Libraries and the Positive Control RNA. The preferred method is to analyze 2 µl of each of the Amplified Indexed Libraries using either an Agilent Bioanalyzer with the Agilent High Sensitivity DNA Kit (Cat.# 5067-4626) or Fragment Analyzer using the High Sensitivity NGS Analysis Kit (Cat.# DNF-473-1000) using the manufacturer’s protocol. Quantifying the PCR products after removing PCR primers is more accurate than quantifying before the primer removal clean-up step.
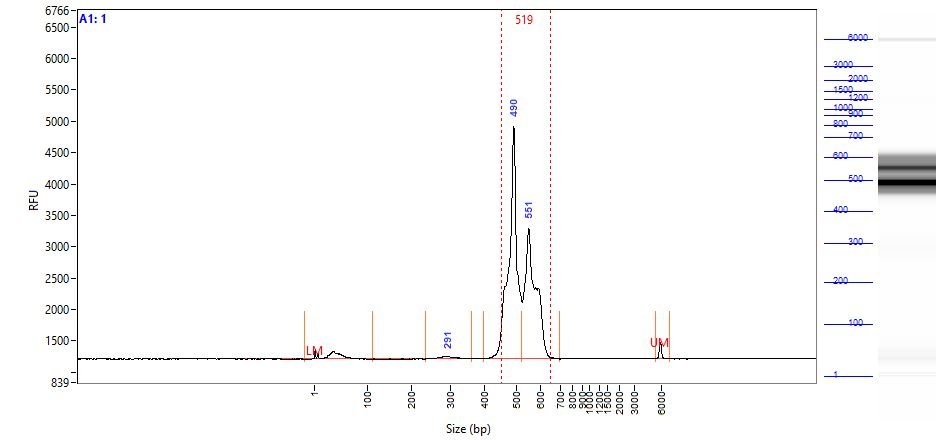
- After primer removal and quantification, use the yield assessment of the Amplified Indexed Libraries as a basis to combine equimolar amounts of each of the Amplified Index Libraries into a single pool for NGS. For example, if the yield of Library 1 is twice that of Library 2, then mix 5 µl of Library 1 with 10 µl of Library 2. To minimize sample-to-sample sequencing variations, combine and load all experimental samples onto one flow cell.
For the DriverMap AIR Assay, which generates a complex pool of amplified TCR/BCR CDR regions (100K-200K from 50 ng of total whole blood, PBMC RNA), refer to the table below for guidelines on how many samples may be combined for different instruments and read depths. Generally, you should aim for 5-10 million reads per AIR sample (starting from 50 ng of PBMC RNA) or at least 20 reads per cDNA molecule. For Illumina instruments, AIR profiling assays require 600-n paired-end reagent kits.
| Instrument | Reads per flow cell | Number of samples for multiplexing per flow cell | Reads per AIR sample |
|---|---|---|---|
| NextSeq 1000/2000 | 100 million (P1 flow cell) | 10-15 | 6-10M reads/sample |
| NextSeq 1000/2000 | 300 million (P2 flow cell) | 30-50 | 6-10M reads/sample |
The multiplexing level may be modified to meet your experimental needs. For example, multiplex fewer samples together to generate more sequencing reads per sample (i.e., more depth) for increased and more sensitive detection of clonotypes present across a broader dynamic range of abundance/expression levels, or, if you are mostly interested only in highly abundant/expressed clonotypes, you can sequence more samples together which will be less expensive.
Need more help with this?
Contact Us

